Kathrin Thüring
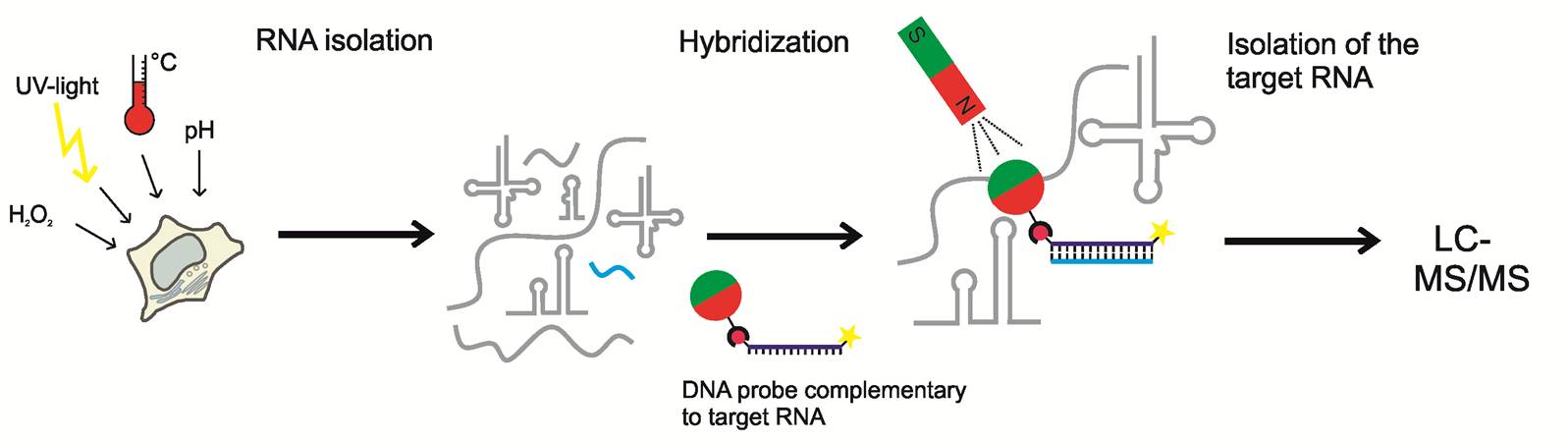
Small non-coding RNAs (ncRNA) fulfill important regulatory functions by modulating gene expression. Like other RNA types, small ncRNAs probably contain nucleoside modifications, and external factors might influence the occurrence of these modifications and thus gene expression. However, the detection of modified nucleosides in small ncRNAs is a challenge because of their low abundance. Hence, first an analytical approach has to be established: the target RNA has to be isolated for modification analysis by highly sensitive LC-MS/MS and the position of detected modification within the target RNA has to be determined. Finally, to investigate the impact of environmental factors on the modification pattern, small ncRNAs from cells subjected to various stress treatments will be analyzed. For the isolation of target RNAs, a hybridization-based approach will be applied, making use of complementary DNA probes immobilized on magnetic beads.